The goal of ggreveal is to make it easy to incrementally show parts of a ggplot. The package offers different ways to split a finished plot into steps.
Examples
Let’s create a plot that to illustrate different ways to reveal
elements. We will use the penguins dataset from the
palmerpenguins package, which contains measurements of
penguins from three different species.
library(palmerpenguins)
library(ggplot2)
penguins <-penguins[!is.na(penguins$sex),]
p1 <- ggplot(penguins,
aes(x = body_mass_g,
y = bill_length_mm,
color = sex)) +
geom_point() +
geom_smooth(method="lm", formula = 'y ~ x', linewidth=1) +
facet_wrap(~species) +
theme_minimal()
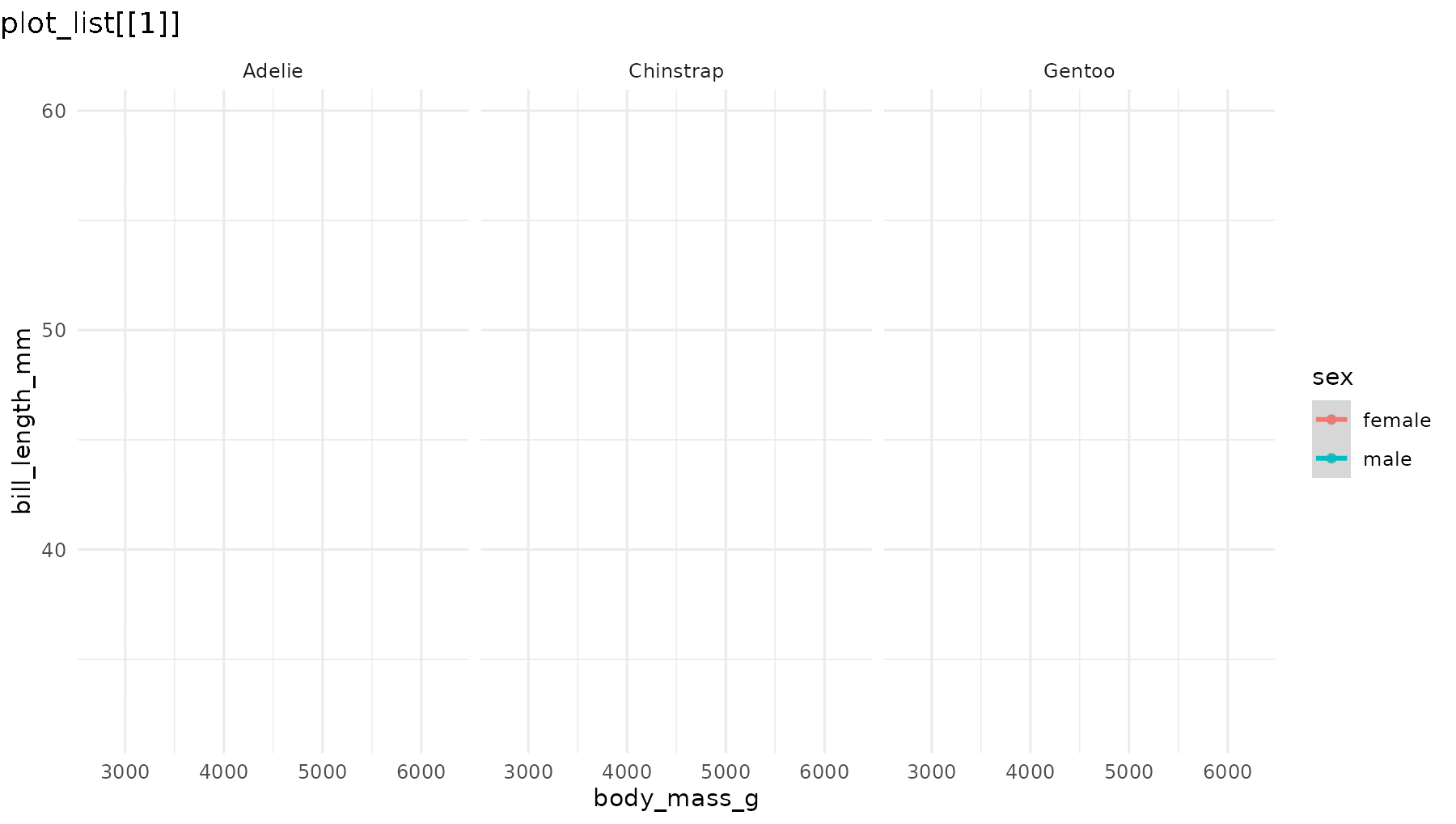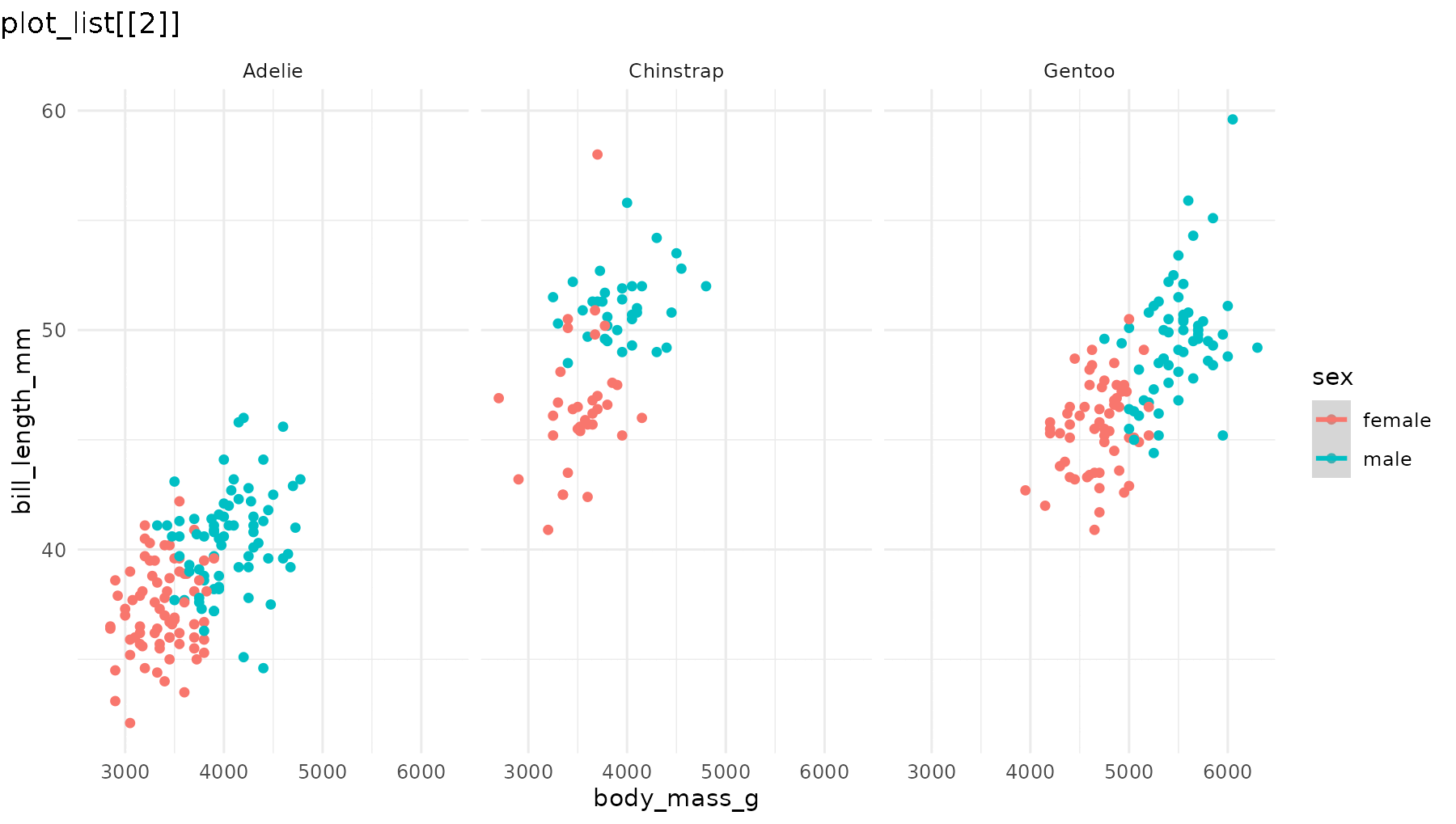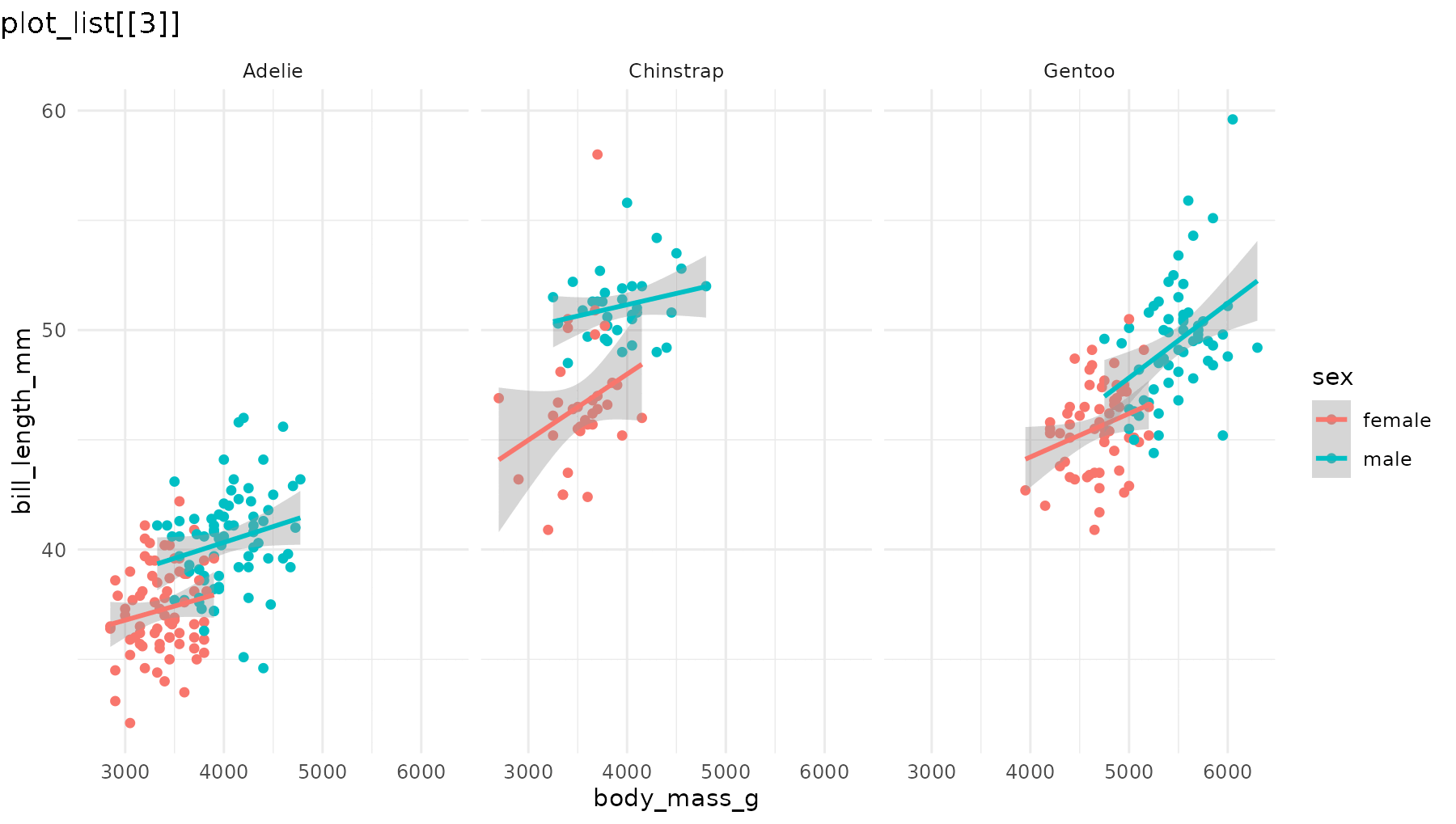
p1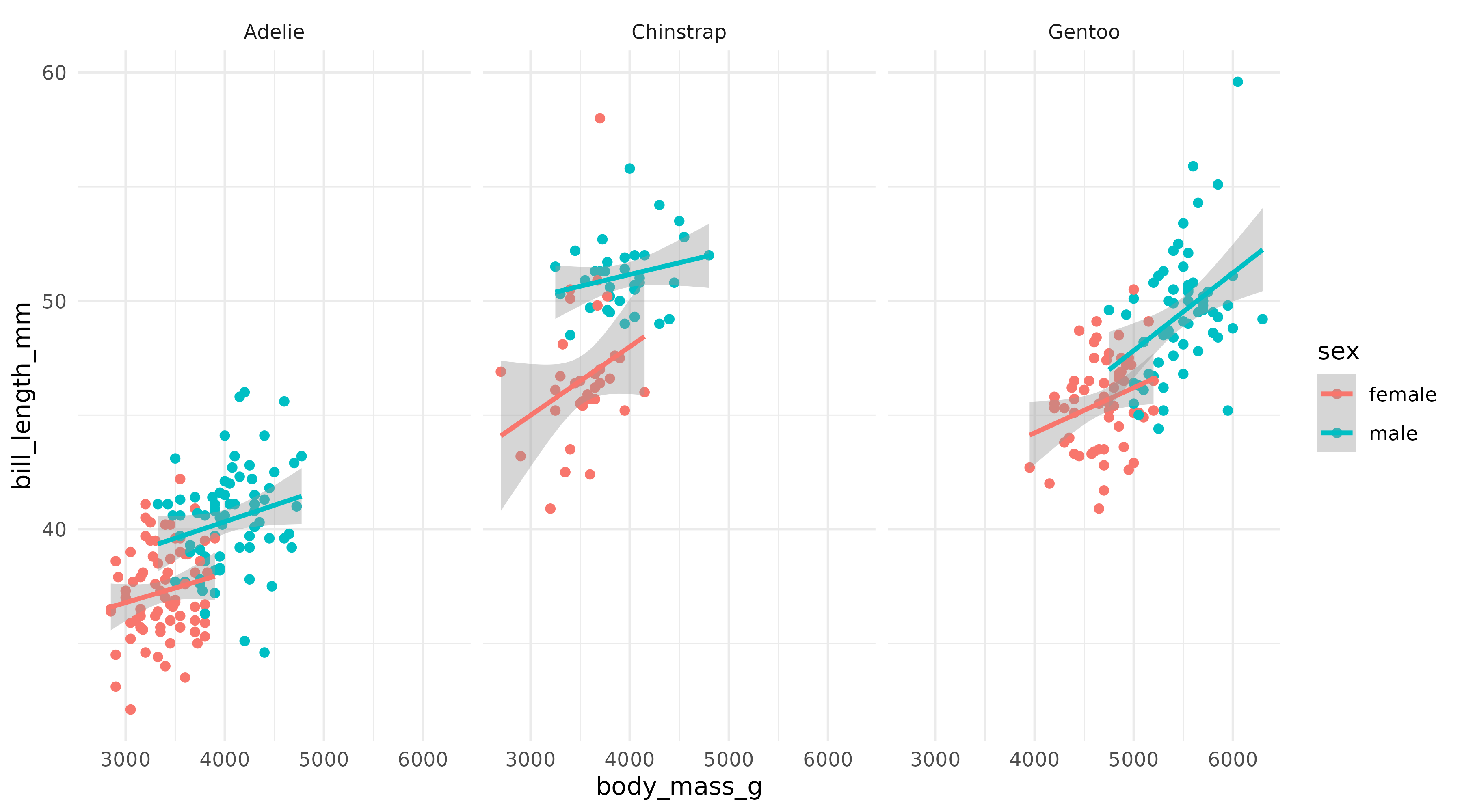
We have a scatter plot of body mass vs bill length, colored by sex and faceted by species. On top of the points, we have linear regression lines for each combination of sex and species.
Reveal by aesthetic
Let’s say that during a presentation we want to highlight the
differences between female and male pennguins. The sex
variable is mapped to the color aesthetic, so we can use it
with reveal_aes().
plot_list <- reveal_aes(p1, aes = "color") plot_list is a list of 3 plots: the first shows only
layout elements (axes, legend, etc) but no data; the second adds the
data for female penguins; and the third adds the data for male penguins,
thus arriving at the original plot.
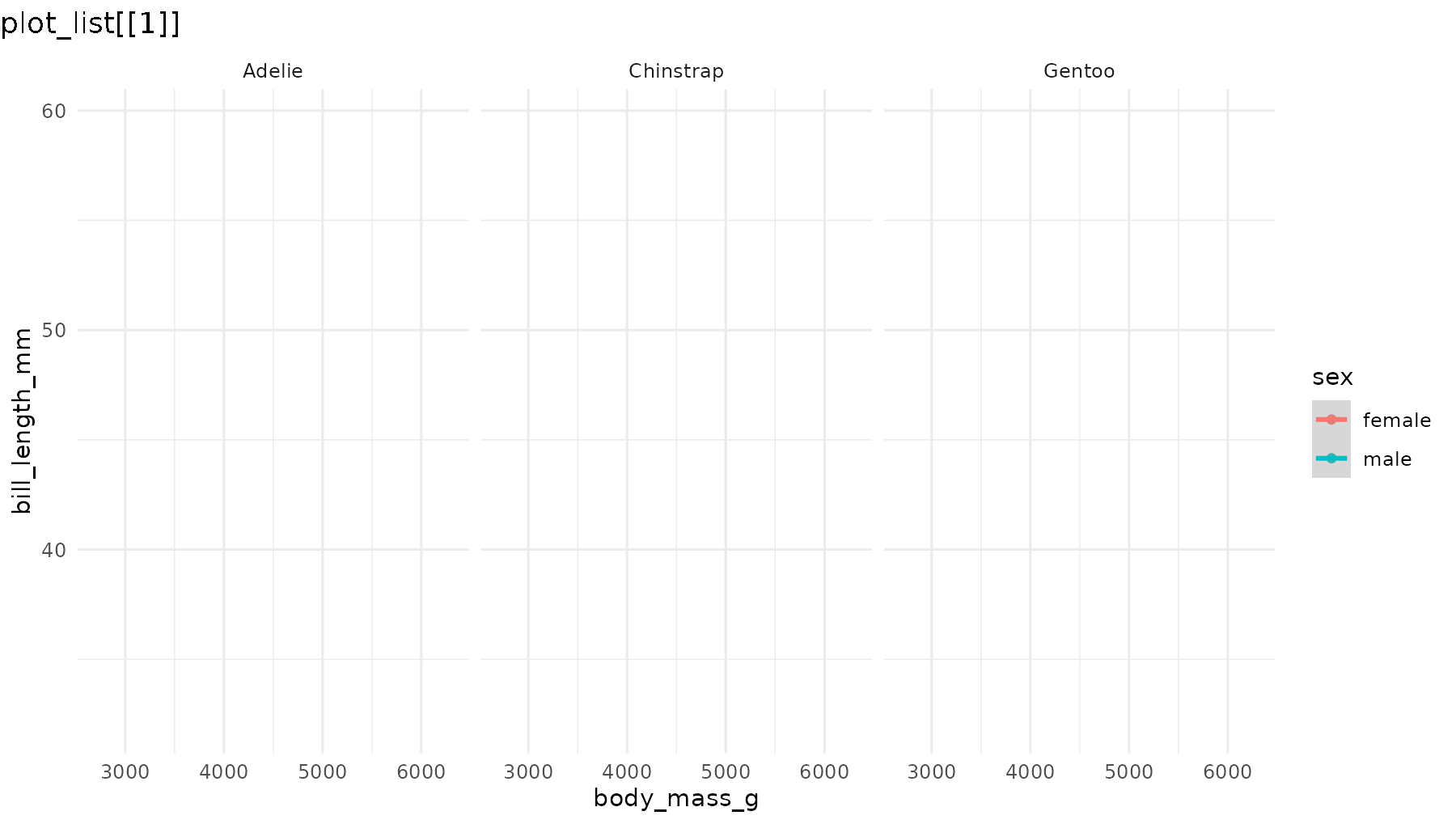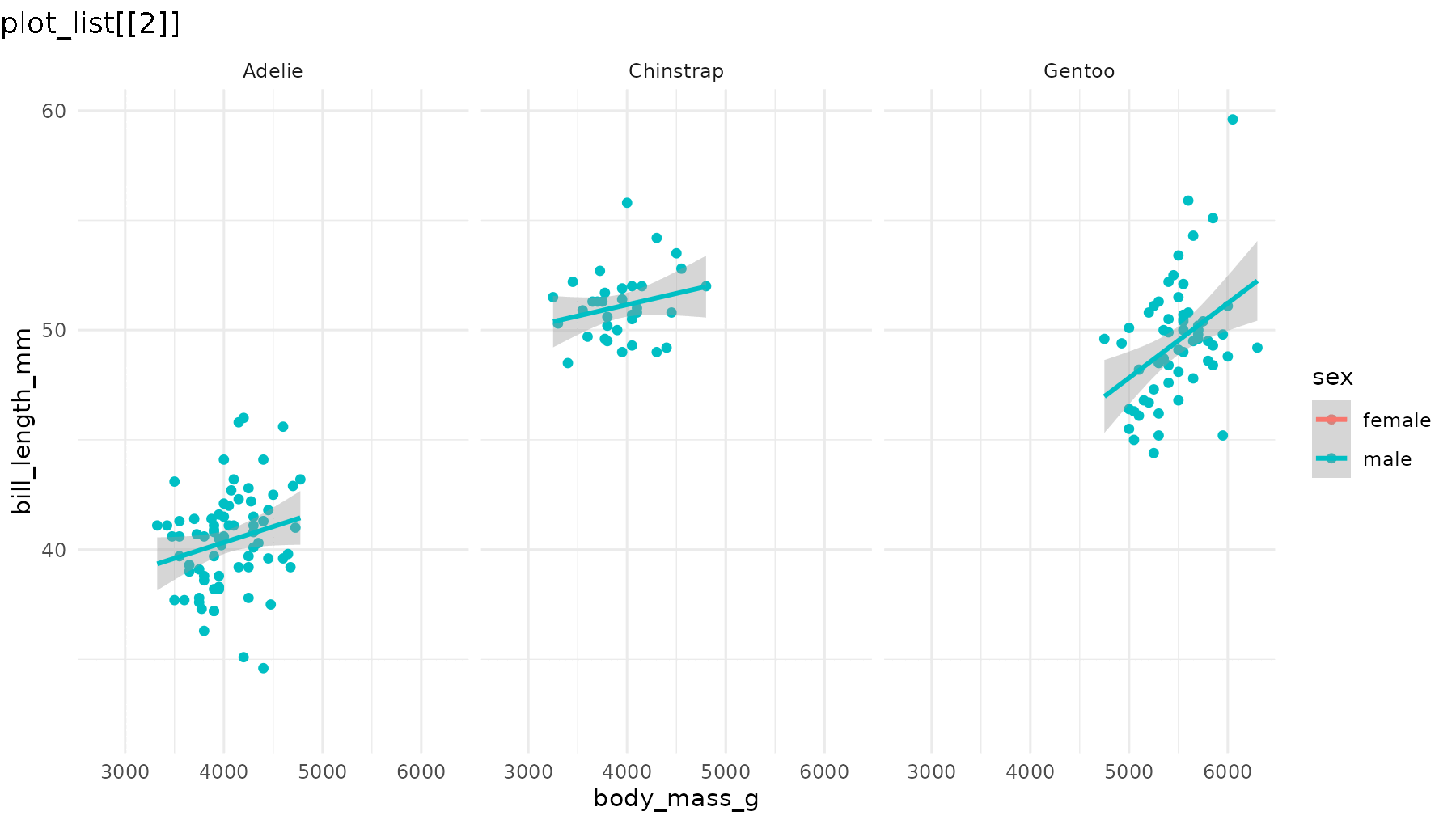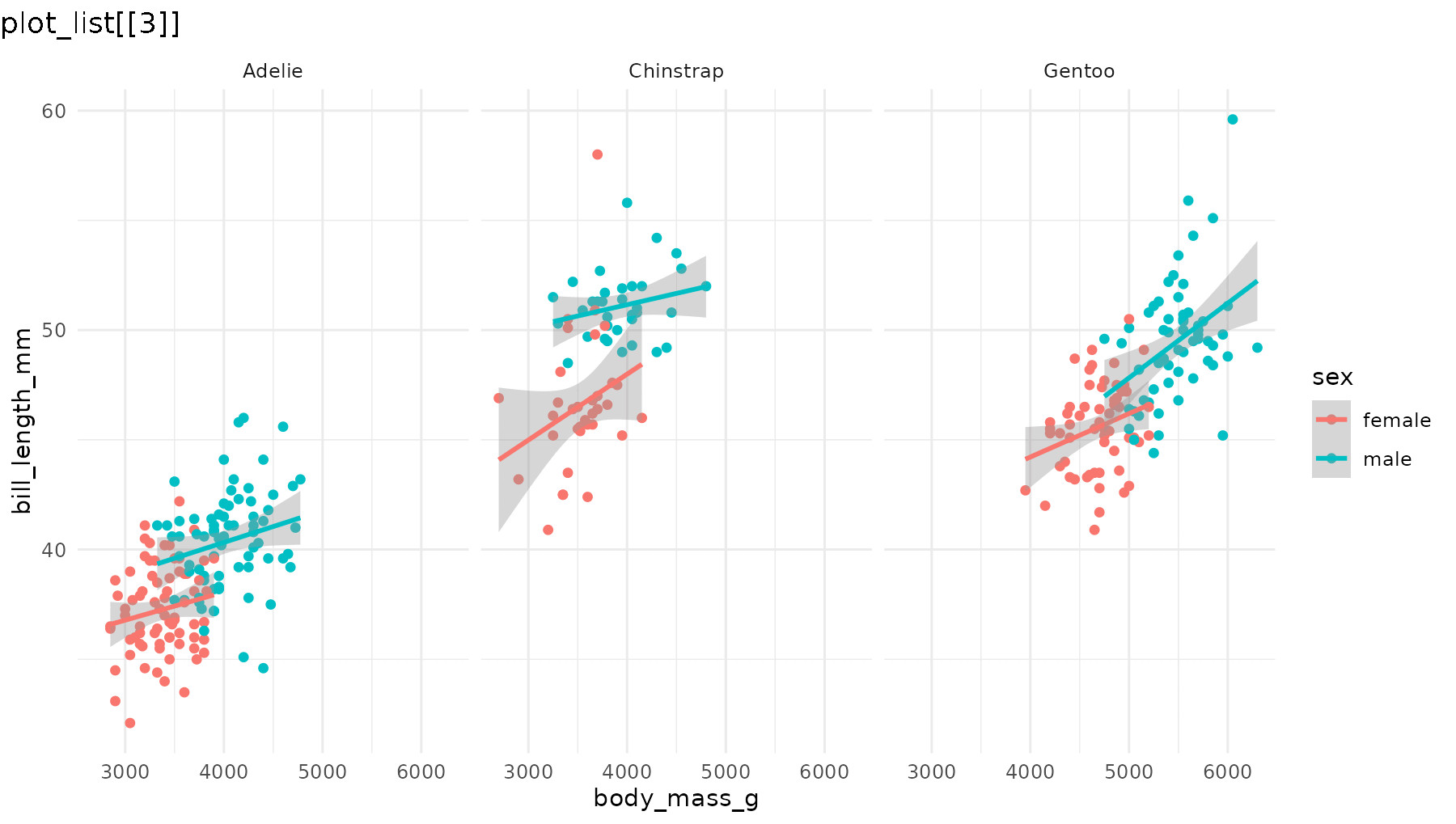
Note: ggreveal does not produce animations. The functions return a list of plots, which are shown here in animated gifs for sake of concision.
The reveal_aes() function is quite flexible and can be
used to reveal pretty much any aesthetic mapping. Suppose we map sex the
shape aesthetic instead: 1
p2 <- ggplot(penguins,
aes(x = body_mass_g,
y = bill_length_mm,
shape= sex)) +
geom_point() +
geom_smooth(method="lm", formula = 'y ~ x', linewidth=1) +
scale_shape_manual(values = c(1,15))
facet_wrap(~species) +
theme_minimal()
plot_list <- reveal_aes(p2, aes = "shape")
plot_list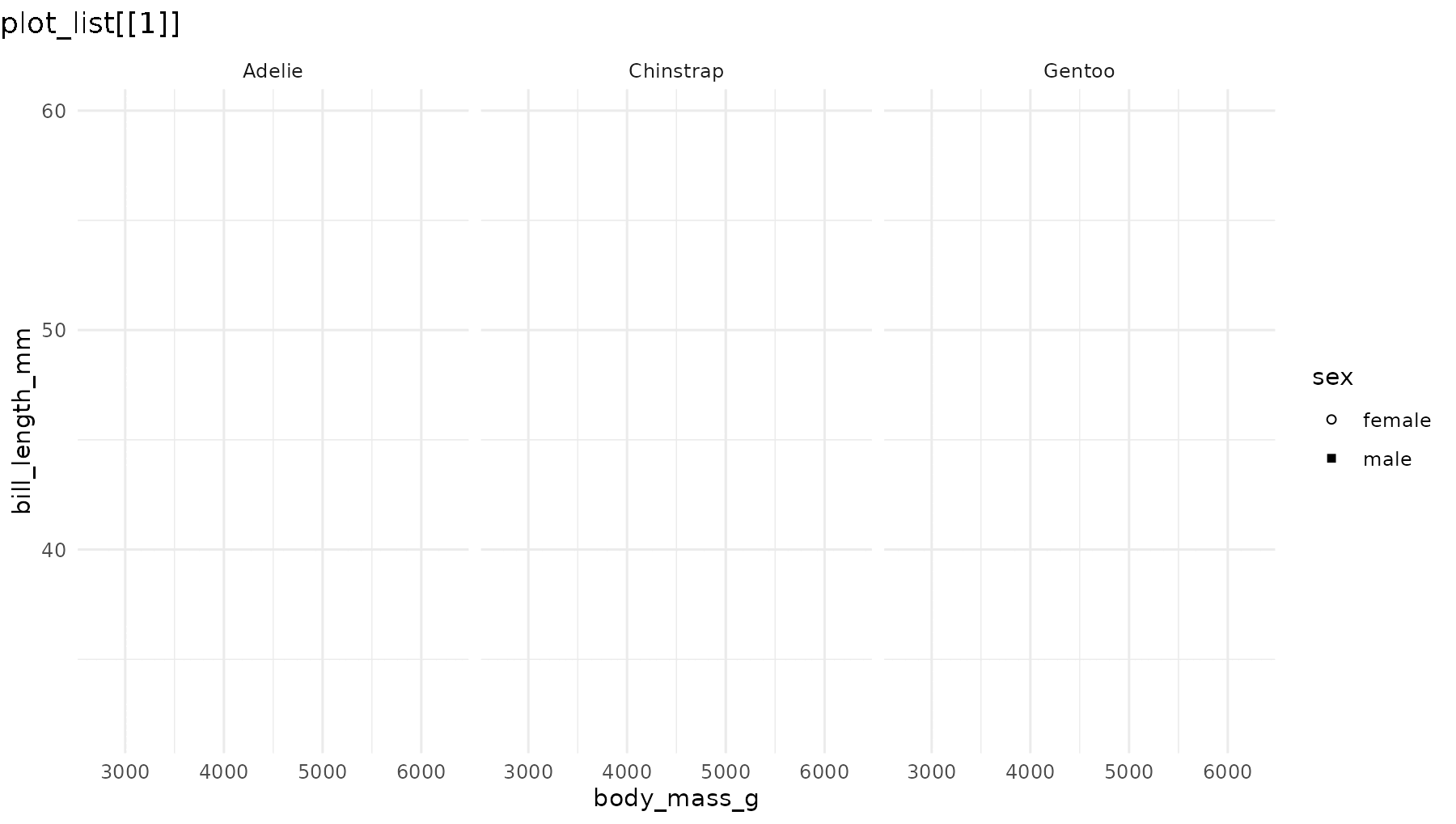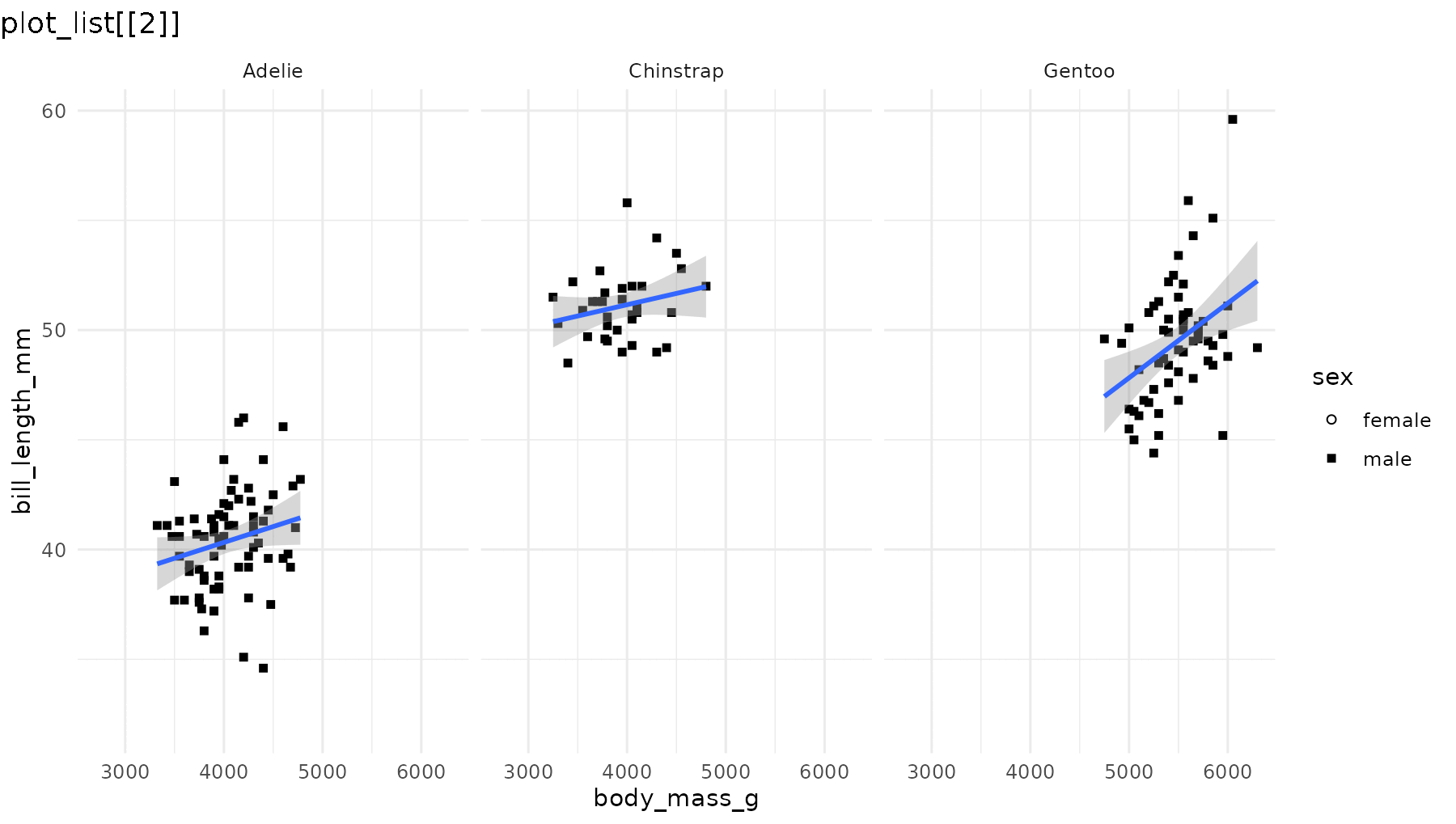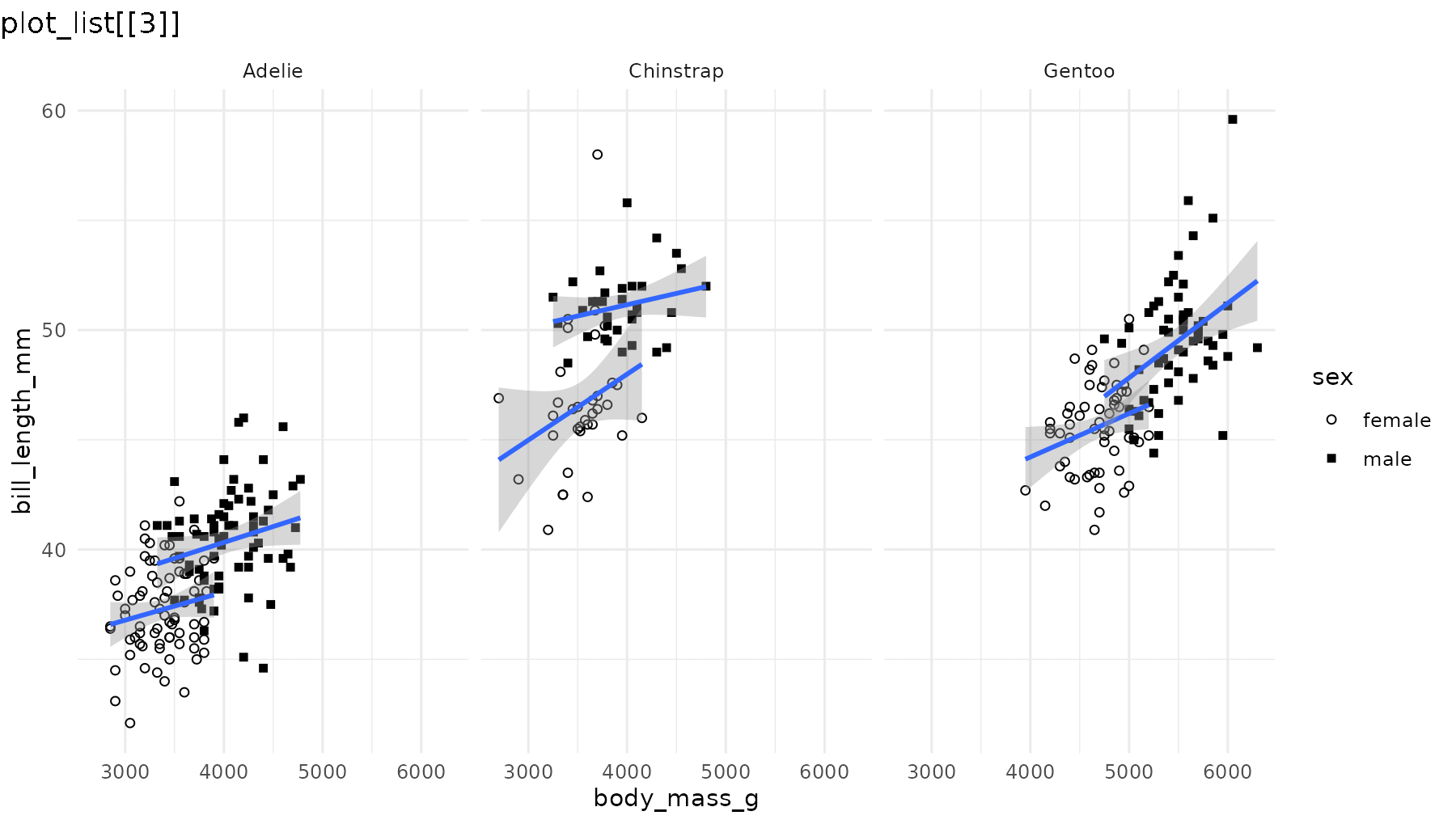
Or say we want to reveal categories in the x axis:
p3 <- ggplot(penguins,
aes(x = sex, y = bill_length_mm )) +
geom_boxplot() +
theme_minimal()
plot_list <- reveal_aes(p3, aes = "x")
plot_list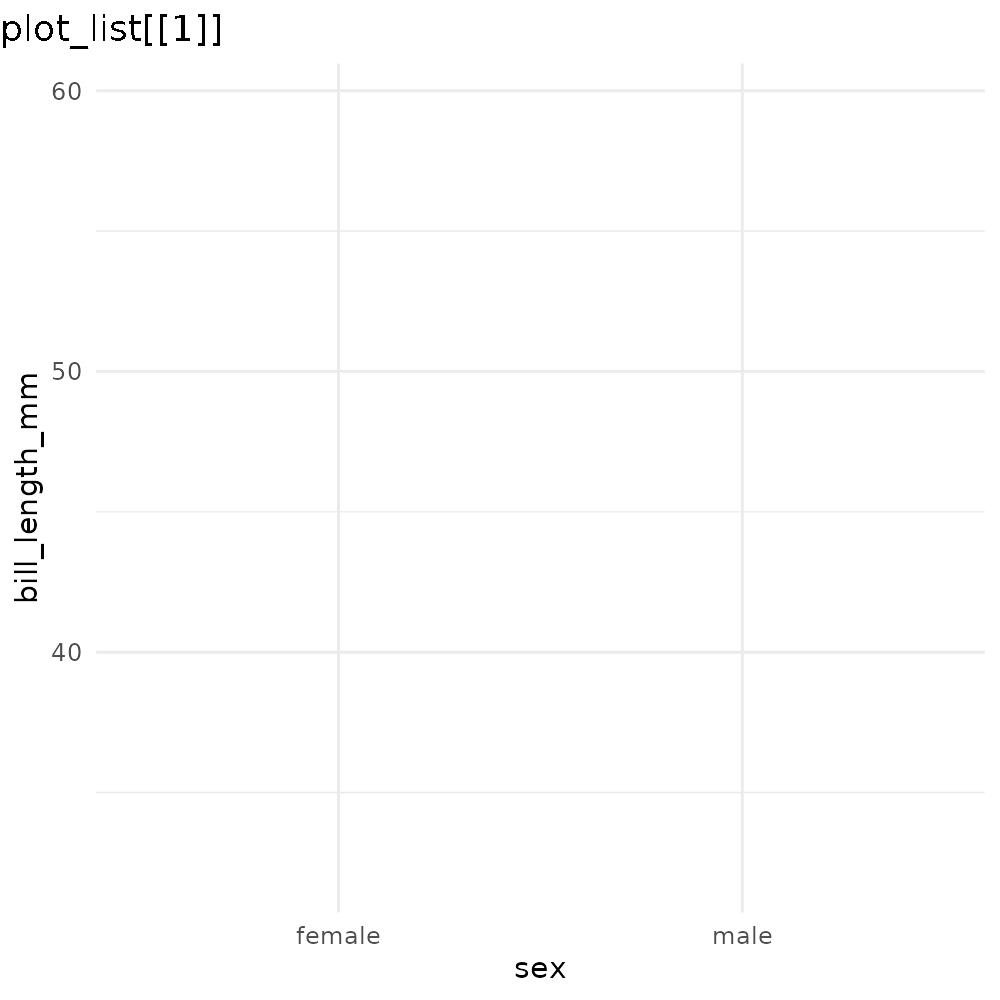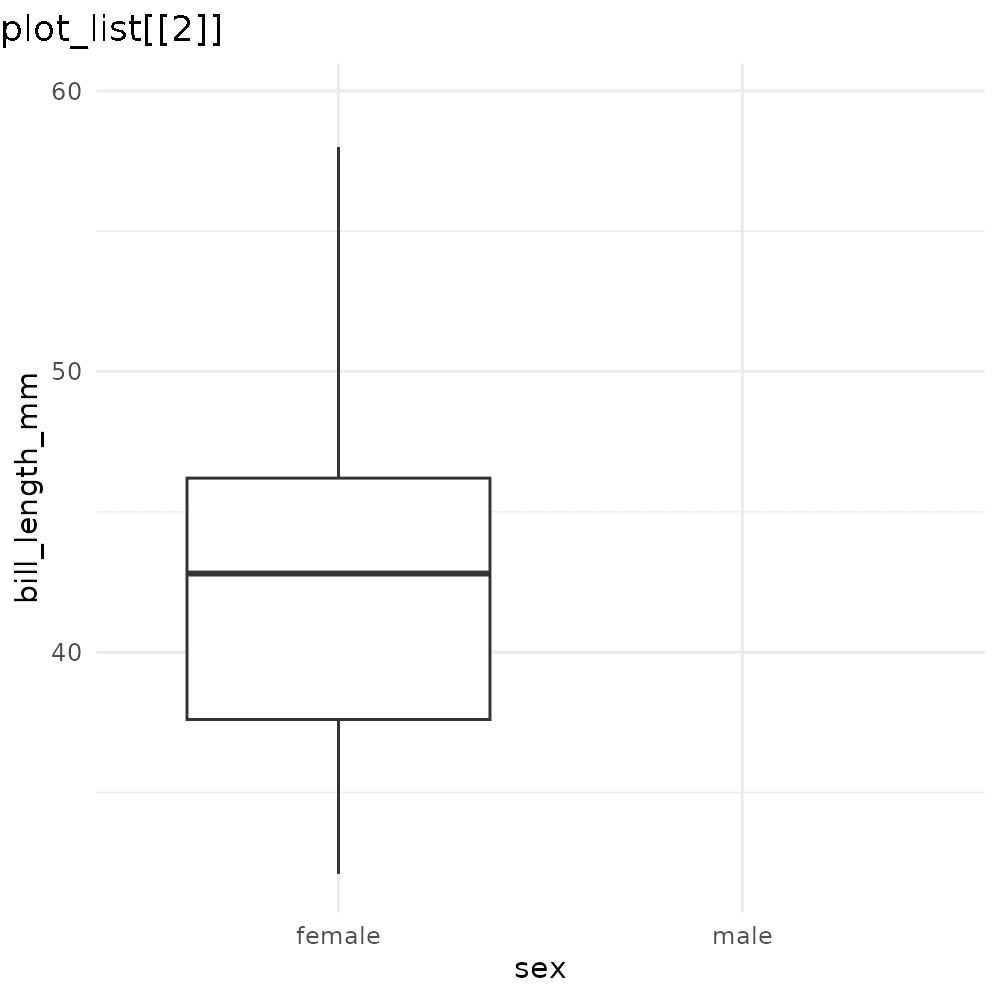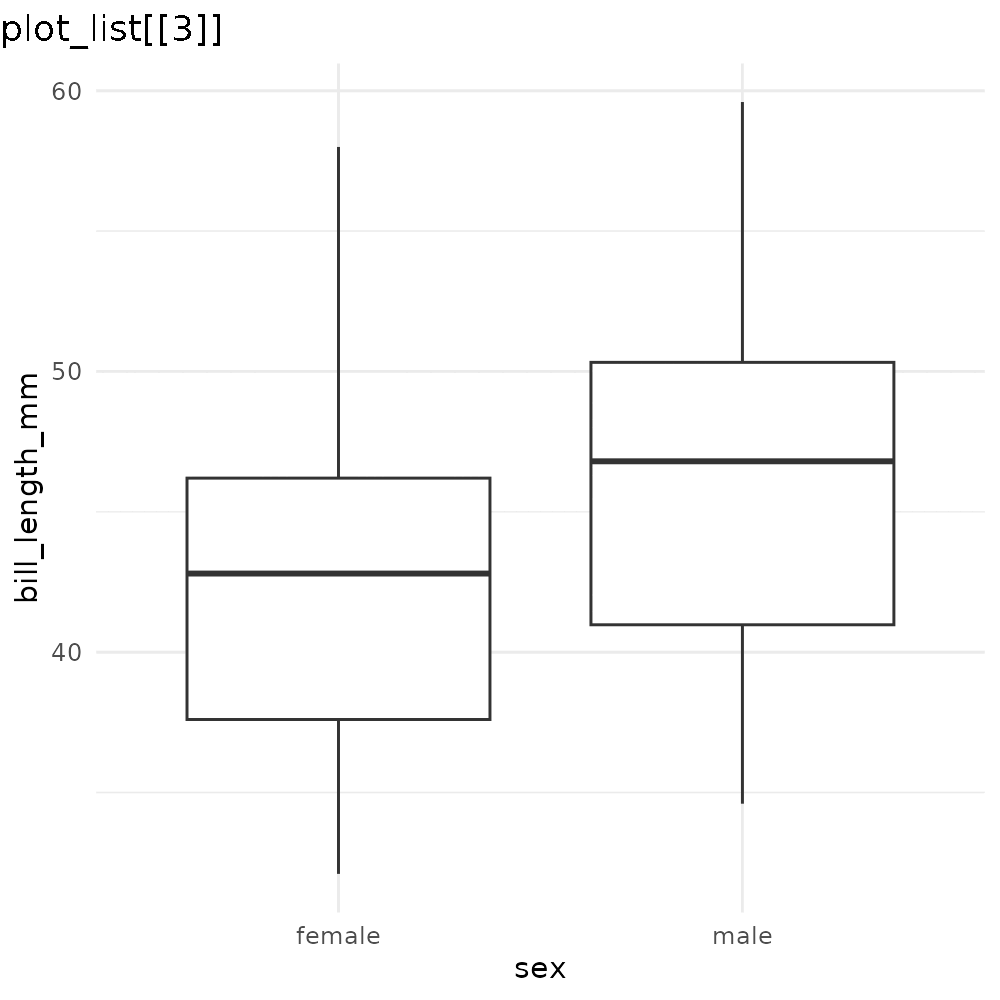
reveal_x() is a shortcut for
reveal_aes(aes = "x"), and similarly for
reveal_y(). Revealing axes works better when the x or y
variable is discrete, or has few values, otherwise you can get a lot of
intermediary plots. If you find yourself needing to incrementally reveal
a large amount of values (e.g. a time series developing over the x
axis), you might want to use gganimate instead.
There is also reveal_groups
Reveal by panel/facet
Returning to our first plot, we might instead want to draw attention
to differences across species, and thus reveal each panel (facet) at a
time with reveal_panels(). Now we get four plots: one
“blank”, then one plot for each of the three species revealed
cumulatively. Again, the last element in the list corresponds to the
original plot:
plot_list <- reveal_panels(p1)
plot_list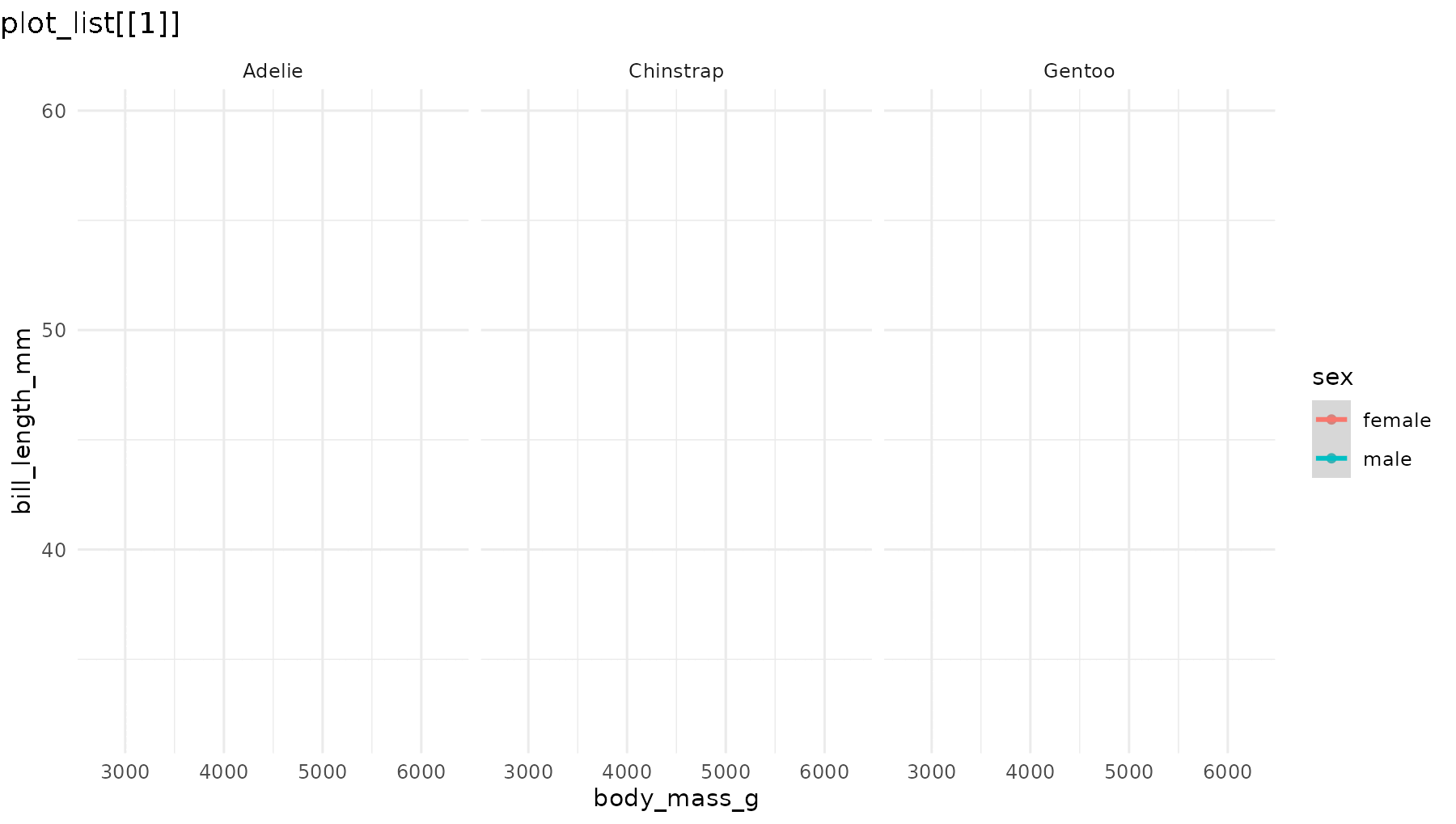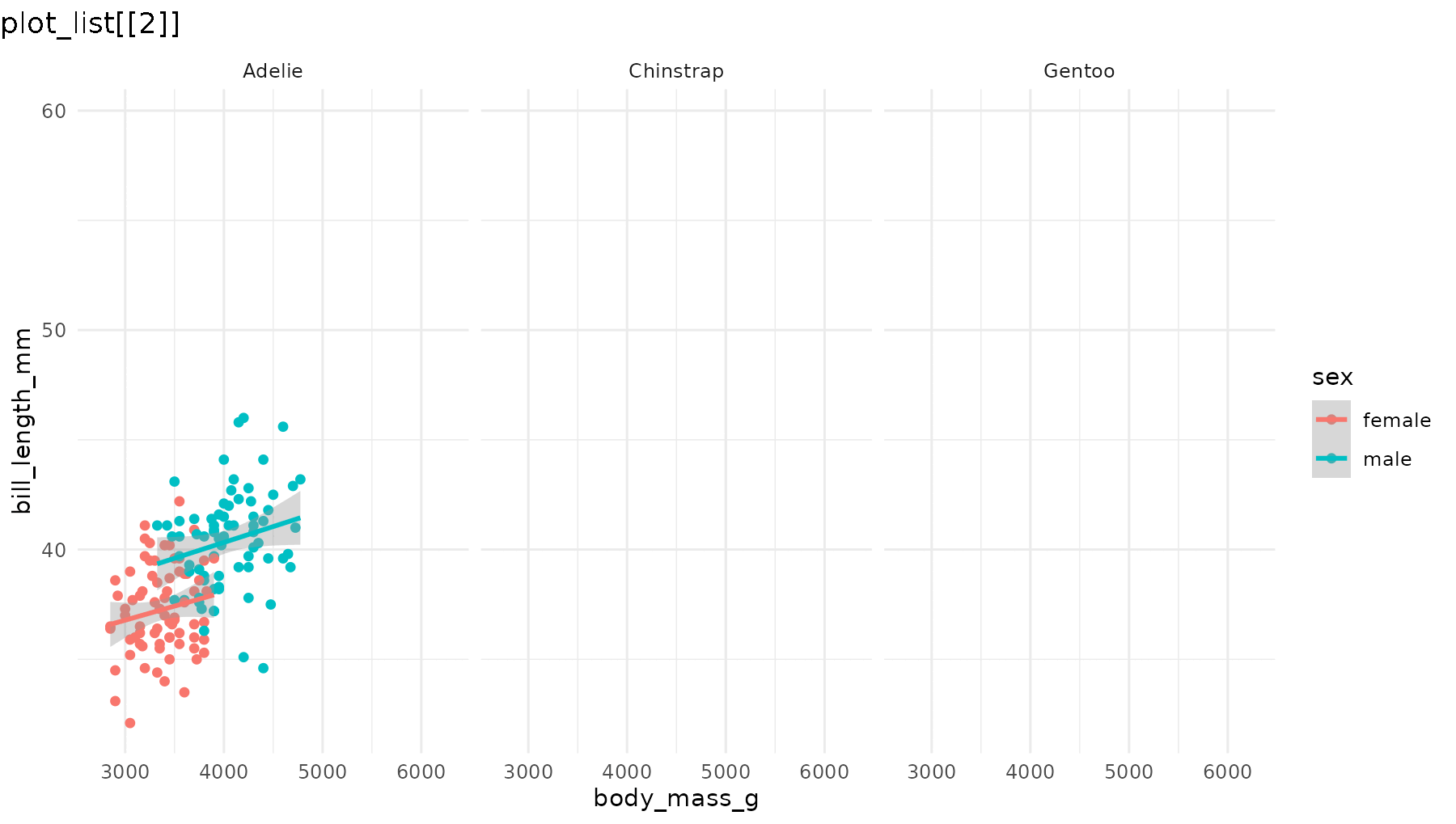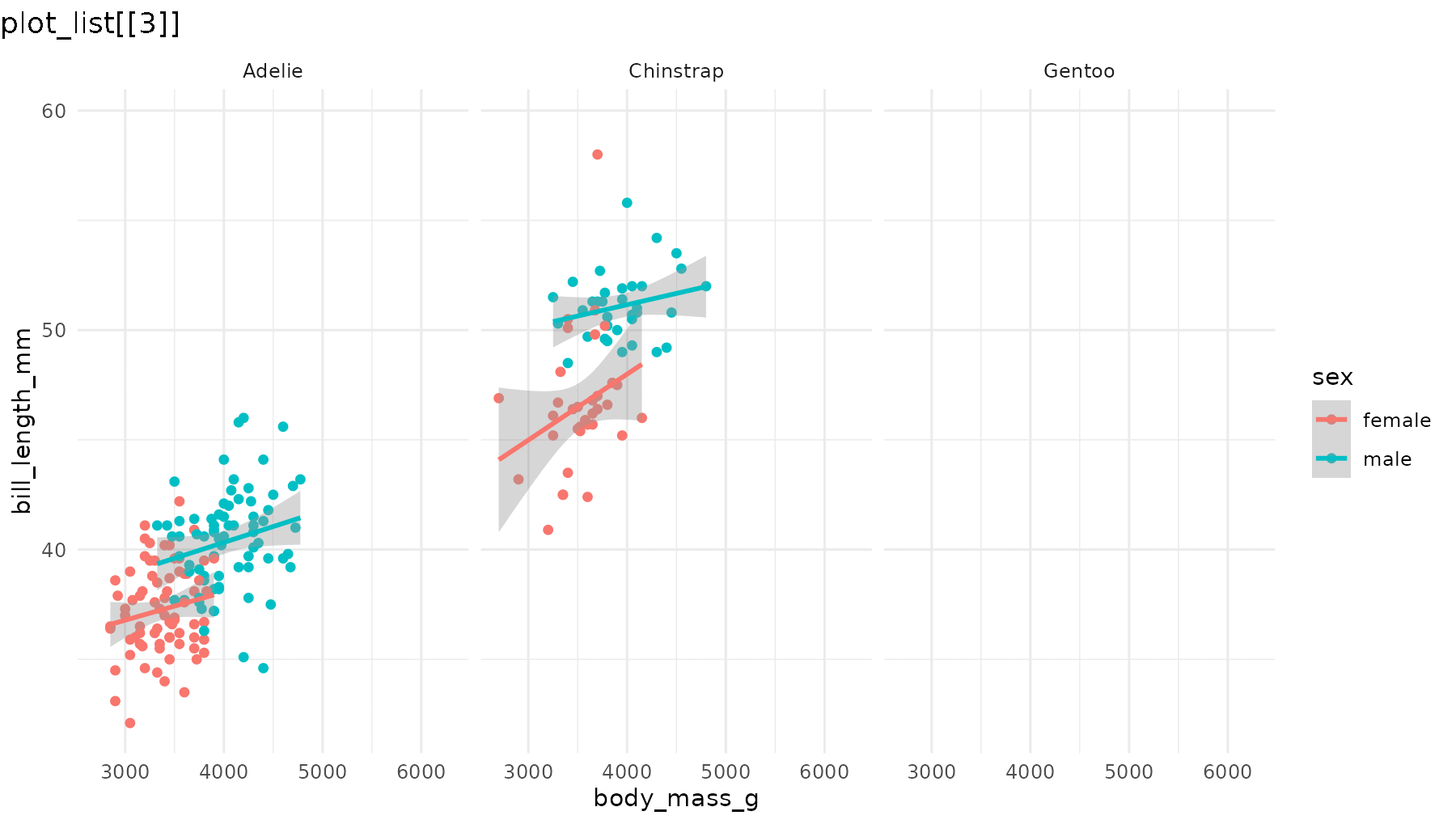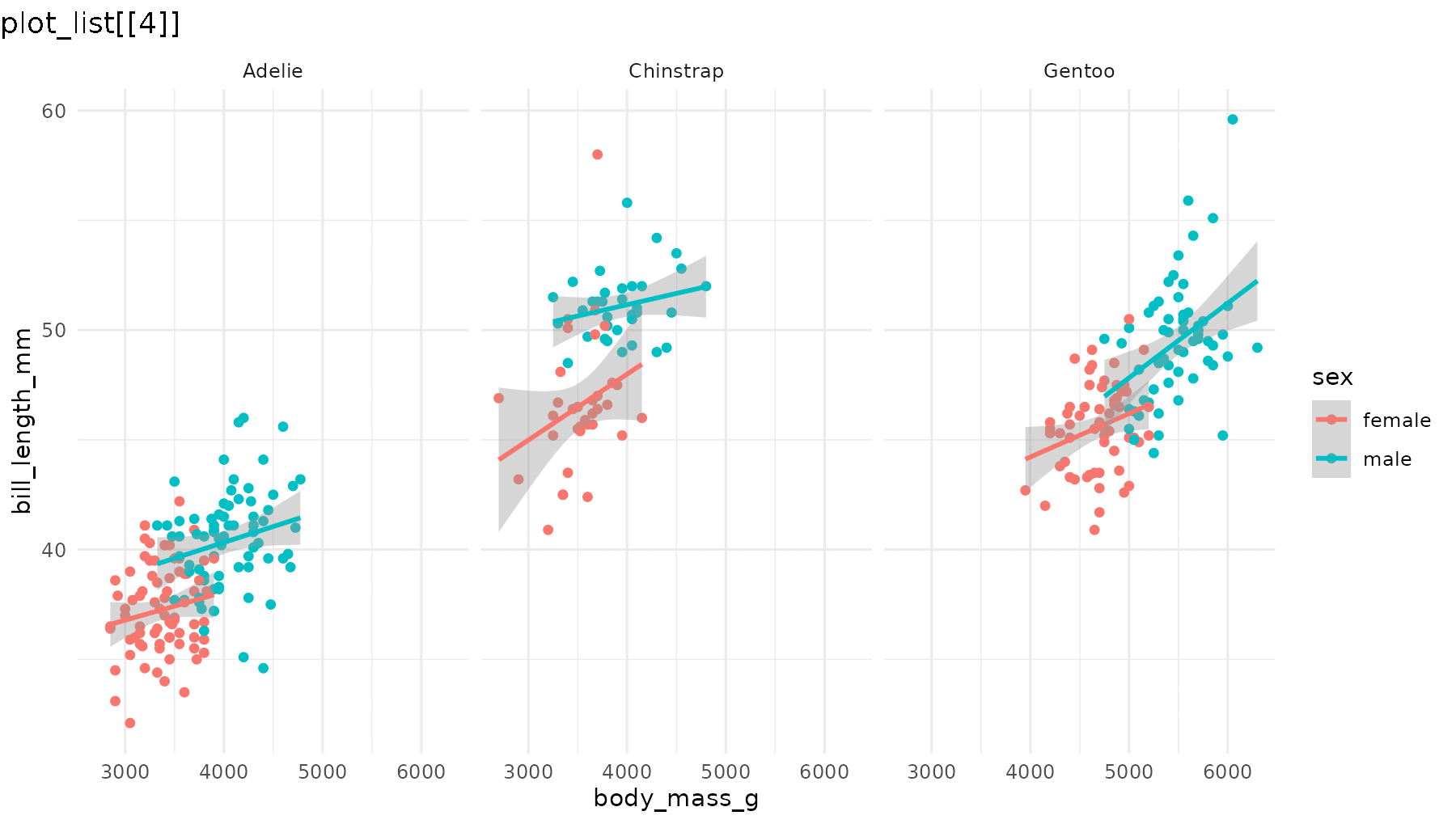
Reveal by layer
Finally, we might want to first show the raw data and then add the
regression lines. These elements are defined by different layers, so we
can use reveal_layers():
plot_list <- reveal_layers(p1)
plot_list